This post is about using OrderBook/PositionBook features as input to simple machine learning models after previous investigation into the relevance of such features.
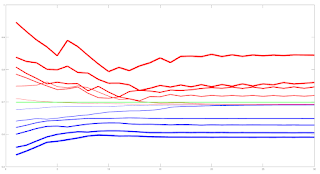
Due to the amount of training data available I decided to look only at a linear model and small neural networks (NN) with a single hidden layer with up to 6 hidden neurons. This choice was motivated by an academic paper I read online about linear models which stated that, as a lower bound, one should have at least 10 training examples for each parameter to be estimated. Other online reading about order flow imbalance (OFI) suggested there is a linear relationship between OFI and price movement. Use of limited size NNs would allow a small amount of non linearity in the relationship. For this investigation I used the Netlab toolbox and Octave. A plot of the learning curves of the classification models tested is shown below. The targets were binary 1/0 for price increases/decreases.
The blue lines show the average training error (y axis) and the red lines show the same average error metric on the held out cross validation data set for each tested model. The thickness of the lines represents the number of neurons in the single hidden layer of the NNs (the thicker the lines, the higher the number of hidden neurons). The horizontal green line shows the error of a generalized linear model (GLM) trained using iteratively reweighted least squares. It can be seen that NN models with 1 and 2 hidden neurons slightly outperform the GLM, with the 2 neuron model having the edge over the 1 neuron model. NN models with 3 or more hidden neurons over fit and underperform the GLM. The NN models were trained using Netlab's functions for Bayesian regularization over the parameters.
Looking at these results it would seem that a 2 neuron NN would be the best choice; however the error differences between the 1 and 2 neuron NNs and GLM are small enough to anticipate that the final classifications (with a basic greater/less than a 0.5 logistic threshold value for long/short) would perhaps be almost identical.
Investigations into this will be the subject of my next post.
The code box below gives the working Octave code for the above.
## load data
##training_data = dlmread( 'raw_netlab_training_features' ) ;
##cv_data = dlmread( 'raw_netlab_cv_features' ) ;
training_data = dlmread( 'netlab_training_features_svd' ) ;
cv_data = dlmread( 'netlab_cv_features_svd' ) ;
training_targets = dlmread( 'netlab_training_targets' ) ;
cv_targets = dlmread( 'netlab_cv_targets' ) ;
kk_loop_record = zeros( 30 , 7 ) ;
for kk = 1 : 30
## first train a glm model as a base comparison
input_dim = size( training_data , 2 ) ; ## Number of inputs.
net_lin = glm( input_dim , 1 , 'logistic' ) ; ## Create a generalized linear model structure.
options = foptions ; ## Sets default parameters for optimisation routines, for compatibility with MATLAB's foptions()
options(1) = 1 ; ## change default value
## OPTIONS(1) is set to 1 to display error values during training. If
## OPTIONS(1) is set to 0, then only warning messages are displayed. If
## OPTIONS(1) is -1, then nothing is displayed.
options(14) = 5 ; ## change default value
## OPTIONS(14) is the maximum number of iterations for the IRLS
## algorithm; default 100.
net_lin = glmtrain( net_lin , options , training_data , training_targets ) ;
## test on cv_data
glm_out = glmfwd( net_lin , cv_data ) ;
## cross-entrophy loss
glm_out_loss = -mean( cv_targets .* log( glm_out ) .+ ( 1 .- cv_targets ) .* log( 1 .- glm_out ) ) ;
kk_loop_record( kk , 7 ) = glm_out_loss ;
## now train an mlp
## Set up vector of options for the optimiser.
nouter = 30 ; ## Number of outer loops.
ninner = 2 ; ## Number of innter loops.
options = foptions ; ## Default options vector.
options( 1 ) = 1 ; ## This provides display of error values.
options( 2 ) = 1.0e-5 ; ## Absolute precision for weights.
options( 3 ) = 1.0e-5 ; ## Precision for objective function.
options( 14 ) = 100 ; ## Number of training cycles in inner loop.
training_learning_curve = zeros( nouter , 6 ) ;
cv_learning_curve = zeros( nouter , 6 ) ;
for jj = 1 : 6
## Set up network parameters.
nin = size( training_data , 2 ) ; ## Number of inputs.
nhidden = jj ; ## Number of hidden units.
nout = 1 ; ## Number of outputs.
alpha = 0.01 ; ## Initial prior hyperparameter.
aw1 = 0.01 ;
ab1 = 0.01 ;
aw2 = 0.01 ;
ab2 = 0.01 ;
## Create and initialize network weight vector.
prior = mlpprior(nin , nhidden , nout , aw1 , ab1 , aw2 , ab2 ) ;
net = mlp( nin , nhidden , nout , 'logistic' , prior ) ;
## Train using scaled conjugate gradients, re-estimating alpha and beta.
for ii = 1 : nouter
## train net
net = netopt( net , options , training_data , training_targets , 'scg' ) ;
train_out = mlpfwd( net , training_data ) ;
## get train error
## mse
##training_learning_curve( ii ) = mean( ( training_targets .- train_out ).^2 ) ;
## cross entropy loss
training_learning_curve( ii , jj ) = -mean( training_targets .* log( train_out ) .+ ( 1 .- training_targets ) .* log( 1 .- train_out ) ) ;
cv_out = mlpfwd( net , cv_data ) ;
## get cv error
## mse
##cv_learning_curve( ii ) = mean( ( cv_targets .- cv_out ).^2 ) ;
## cross entropy loss
cv_learning_curve( ii , jj ) = -mean( cv_targets .* log( cv_out ) .+ ( 1 .- cv_targets ) .* log( 1 .- cv_out ) ) ;
## now update hyperparameters based on evidence
[ net , gamma ] = evidence( net , training_data , training_targets , ninner ) ;
## fprintf( 1 , '\nRe-estimation cycle ##d:\n' , ii ) ;
## disp( [ ' alpha = ' , num2str( net.alpha' ) ] ) ;
## fprintf( 1 , ' gamma = %8.5f\n\n' , gamma ) ;
## disp(' ')
## disp('Press any key to continue.')
##pause;
endfor ## ii loop
endfor ## jj loop
kk_loop_record( kk , 1 : 6 ) = cv_learning_curve( end , : ) ;
endfor ## kk loop
plot( training_learning_curve(:,1) , 'b' , 'linewidth' , 1 , cv_learning_curve(:,1) , 'r' , 'linewidth' , 1 , ...
training_learning_curve(:,2) , 'b' , 'linewidth' , 2 , cv_learning_curve(:,2) , 'r' , 'linewidth' , 2 , ...
training_learning_curve(:,3) , 'b' , 'linewidth' , 3 , cv_learning_curve(:,3) , 'r' , 'linewidth' , 3 , ...
training_learning_curve(:,4) , 'b' , 'linewidth' , 4 , cv_learning_curve(:,4) , 'r' , 'linewidth' , 4 , ...
training_learning_curve(:,5) , 'b' , 'linewidth' , 5 , cv_learning_curve(:,5) , 'r' , 'linewidth' , 5 , ...
training_learning_curve(:,6) , 'b' , 'linewidth' , 6 , cv_learning_curve(:,6) , 'r' , 'linewidth' , 6 , ...
ones( size( training_learning_curve , 1 ) , 1 ).*glm_out_loss , 'g' , 'linewidth', 2 ) ;
## >> mean(kk_loop_record)
## ans =
##
## 0.6928 0.6927 0.7261 0.7509 0.7821 0.8112 0.6990
## >> std(kk_loop_record)
## ans =
##
## 8.5241e-06 7.2869e-06 1.2999e-02 1.5285e-02 2.5769e-02 2.6844e-02 2.2584e-16